I’m always glad whenever I see a microbiology theme breach the moat of the NYT and make it on to their opinion page. But I usually end up disappointed once I actually read it.
That’s the case with this call to biobank the world’s human gut microbiome. It’s far from the worst idea published this week (that prize goes to a recipe for Cheeto Turkey), but it is lazy and superficial and so I feel called to dump on it. Which maybe is not the right term since we are talking about stool samples, but there we are.
The author rightly observes that the gut microbiome diversity of city-dwellers is about half that of hunter-gatherers, and worries that as the world becomes more urbanized – and thus subject to antibiotic exposure, C-sections and other microbe-unfriendly practices – that many potentially useful or beneficial bacterial species will go extinct. In this she echoes an argument made by some leading microbiomicists.
All true enough, I suppose. But no argument is better than its premises, and unstated premises are a sure sign of a weak argument. The unstated premise here is that it is possible for bacteria to go extinct. And that species extinction matters. I challenge both.
Killing all the bacteria in an environment otherwise conducive to life is probably impossible. Antibiotics won’t do it; they won’t even come close. A really powerful antibiotic in test-tube conditions will kill, at most, 99.99999% of the bugs exposed to it. But there is always a population of “persister” bugs that are not actively resistant yet are in a metabolic state that renders them unaffected by the treatment. They may be 0.00001% of the population, but a small fraction of a large number is still a large number. Given adequate nutrients, those few thousand bugs will become millions and then billions again within a few hours. What is true of antibiotics is true for all other kinds of insults to bacteria. They may be massacred but they will not be eliminated.
Flowers and animals go extinct once their populations become sparse because they can’t find mates. Bacteria suffer no such liability. So long as a single cell survives, their populations will quickly rebound if given a chance.
Just because we don’t detect certain bacteria in a stool sample (which is not necessarily representative of the gut microbiome anyway) doesn’t mean they are not there. I suspect if the the average NYT editor began eating more grubs, roots and leaves, and less ramen and kale, their microbiome would become far more diverse within weeks.
But it still probably would not have all the species found in the gut of a typical forest-dweller. I’m doubtful that this matters, however.
Bacteria don’t really have species. Whereas humans (who are aberrantly inbred) share 99.5% of their genes, the comparable number for a bacterial species is lower. Far lower.
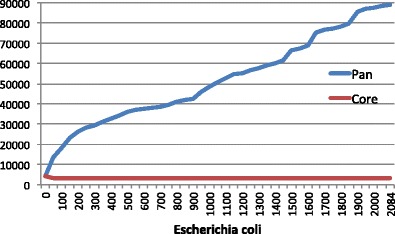
Here’s a summary of the genes found in something that humans call a bacterial species (E. coli) but in reality is something else entirely:
From Insights from 20 years of bacterial genome sequencing
About 3000 genes (the red line) out of the 5000 found in any individual cell comprise the E. coli core genome. Probably over 100,000 genes populate its pan genome. So two strains of E coli may share no more than 60% of their genes. That’s about the same number of genes you share with your houseplants. Using the word “species” to describe both humans and E coli stretches its meaning beyond any usefulness.
We should think of bacteria not as we do of animal or plant species, but as dynamic consortia of genes. And what it true of individual bacteria is especially true of microbiomes. So long as a microbiome collectively encodes a sufficient number and diversity of genes – its metagenome – the actual number and diversity of its species matters little. Many functionally equivalent microbiomes can be constructed for any given metagenome. It is function, not labels, that we need to conserve.
Conserving bacterial species requires their cultivation, which is not a trivial task, even for gut microbes. Storing their DNA is far simpler and serves the same purpose. DNA is easy to extract, chemically very stable and extraordinarily information dense. The technology to reconstruct bacteria from genes is well-advanced.
So we should do this, but not waste time on trying to culture thousands of different bacterial species, a project that will take decades. A couple of grad students and a few freezers are all that is required to preserve the world’s collective human gut metagenome. We should be able to manage that.