Every new discipline goes through an arc of development. First there are the easy discoveries made possible by new technologies or new understandings. Alfred Hershey, who with Martha Chase helped prove that DNA is the carrier of genetic information, once remarked that scientific happiness is “…to have one experiment that works, and keep doing it all the time”. This state of affairs has been known as “Hershey Heaven” ever since.
Microbiomics has been living in Hershey Heaven since Norm Pace and colleagues pioneered the use of DNA sequencing to explore microbial communities. The basic paradigm is simple: find an interesting or unusual habitat, sequence some or all of the DNA within, discover new and remarkable organisms and communities. Publish and advance your career.
Nowhere has this paradigm been applied more vigorously than in the study of the gut microbiome. The samples – poop – are cheap and easy to collect. You find two groups of people who differ in some significant respect (sick vs well, vegetarian vs meat-eater, male vs female, hunter-gatherer vs city-dweller, East vs West, etc, etc, etc), sequence the bugs in their poop, apply a UniFrac analysis , and you are bound to find some interesting correlations and observations. It is an experiment that simply cannot fail.
But career success is not the same as advancing knowledge. Despite mounds of data, no one can say what constitutes a healthy gut microbiome (although pathological ones are easier to recognize). No one can say what changes in a gut microbiome might advance health, which changes might trigger disease, which are irrelevant. These uncertainties are greatest at the level of the individual. For every example, there is a counter example.
In such cases, one naturally suspects that scientists have been looking under the lamppost, restricting their searches to a pool of light that illuminates but also constrains.
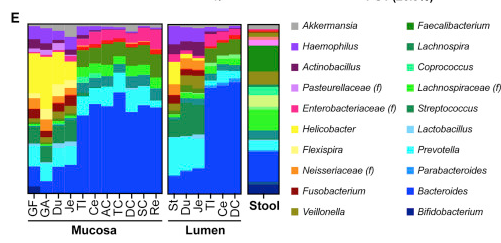
A group of Israeli researchers have now stepped into the dark of the gut microbiome. Working in both mice and humans, they sampled not only poop, but lumen contents and mucosal epithelium at various locations throughout the gut.
They found that poop microbiome composition correlates only modestly with microbiome composition in the gut itself, and that there is significant variation along the course of the gut.
Microbiome composition from top to bottom in the gut, reading from left to right. GF-Je are the stomach and small intestine, TI-Re are the large intestine. . From Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features
Not surprisingly, the stomach and upper intestines show a different pattern of colonization than the lower intestines. Both are different from stool. Bifidobacteria, abundant in stool, and considered among the most beneficial bacteria, are nearly absent in the intestinal mucosa and lumen. Bacteroides are far more prevalent than would be expected from stool profiles.
That is a key result, and would suffice to make this a significant paper. But the authors dug much deeper. There’s a good argument to be made that the consortia of species in the gut microbiome matters far less than its consortia of genes – the metagenome. After all, species are a fairly meaningless, or at least malleable, concept in bacteria. They swap genes all the time anyway, and so the genes being expressed tell us much more about functionality than does a catalog of species.
And here, at the level of metagenome, there is also substantial discordance between stool samples and the gut lumen:
The authors could have stopped here and had a very fine and substantial publication. But this work was largely a setup for an investigation of the impact of probiotics on both mouse and human gut microbiomes.
Probiotics are a big business. Sales are over $2B/year in the US and growing by 12% annually. They are regulated by the FDA as nutritional supplements rather than as therapeutics. This means that manufacturers don’t have to prove (nor can they make) any claims for clinical benefit. But, not surprisingly, plenty of folks are ready to make such claims anyway (and no, I am not going to link to any of their websites).
There is fairly decent evidence for benefits of probiotics in treating antibiotic-associated diarrhea. Depression might also be responsive to probiotics — their effect is comparable to that of antidepressants, which is to say, not all that great.
But most studies, particularly on healthy individuals, show little benefit. Effects on BMI or lipid profiles or insulin sensitivity show small effects or none at all.
No one who thinks much about microbial ecology will be surprised at this. A defining feature of bacteria is their ability to multiply rapidly to saturate every available ecological niche. A rich, stable, moderate environment such as the gut is not very likely to have room for new bacteria absent some stressor (such as antibiotics) that clears out the current inhabitants. This phenomenon is known as colonization resistance – the inability of introduced bacteria to displace established strains.
The Israeli group carefully showed that probiotic bacteria do indeed make it through the stomach and into the gut, but that they mostly get pooped out. On the whole, there was little evidence that of an increase in the population of the probiotic bacteria compared to levels before and after probiotic administration:
That’s at the group level. Some individuals did show establishment of particular strains. The researchers showed that acceptance or rejection of probiotic strains was largely mediated by the existing microbiome. They transplanted individual human microbiomes into germ-free mice, fed them probiotics, and found similar patterns of establishment of probiotics in the animals. Analysis of RNA expression in human hosts also showed differences between subjects that were resistant vs permissive with respect to probiotic colonization.
The takeaways here are that:
- Actual gut microbiome composition varies significantly from stool microbiome composition, and varies both longitudinally and laterally (ie, mucosa vs lumen).
- Probiotics, on the whole, do little to alter microbiome composition in healthy adults
- However, individuals do vary in their susceptibility to colonization with probiotic strains.
- This variation is driven by both existing microbiome composition and host gene expression patterns
None of this is hugely surprising. But it raises the bar significantly for future studies. The “collect poop and sequence” paradigm – the current Hershey Heaven for microbiome studies – is no longer good enough. If microbiomics is going to be applied to improve human health, we have to develop better tools for sampling and describing the microbiomes of individuals. Microbiome-based therapies cannot possibly be one-size-fits-all.
The downside of this is that microbiome studies are going to become more difficult and expensive, at least for a while. The upside is that microbiomic interventions will be smarter, and thus far more likely to succeed. That’s progress.


Interesting, I did not realize the composition is that different between stool and lumen samples.
https://www.nature.com/articles/s41586-018-0616-y
This recent paper shows how probiotics could still improve human health without colonization. You might be interested.