There’s a good chance that we will never be able to use CRISPR in otherwise healthy human beings. Not safely and ethically, anyway.
Let’s assume that all the obvious technical problems are solved – that we learn which genes to modify and how, and that we learn how to deliver CRISPR-Cas complexes safely and efficiently to human tissues, including fertilized eggs. And let’s further stipulate that society reaches a consensus that making people taller or stronger or more attractive is just fine, or at least is nobody else’s business (so long as parents do not use CRISPR to create children that hunt truffles in the forest like pigs etc).
These sort of genetic modifications will still be unethical because they will be unsafe.
Off-target effects – the modification of an unintended gene – are a major limiting factor in CRISPR application today, and may never be controlled enough to use in healthy human tissues.
The specificity of CRISPR depends on the accuracy of base-pairing interactions between its guide RNA and target DNA.
The thermodynamics of basepairing are such that there is not a huge difference in stability between perfect matches and single mismatches. Generally speaking, this difference will result in only a 5- or 10-fold preference for the perfect match sequence over the single mismatch sequence.
And while there is only one perfect match sequence (usually) in a genome, there are many single mismatch sequences. There are three potential mismatches at every position in the guide sequence, which can be 20 nucleotides long, yielding 60 potential single base mismatch sequences. Each one of these forms complexes that are 10-fold less stable than the target sequences, but there are 60 of them. In this (grossly oversimplified) scenario, you’d expect 6 off-target editing reactions for every on-target reaction.
Dan Herschlag wrote up a rigorous treatment of DNA target binding specificity that you can read here.
Quantifying off-target reactions is difficult, but there is no question that they are common, just as expected.
The inherent lack of specificity of DNA and RNA basepairing is not a new problem. It is always an issue in PCR, for instance. But long before CRISPR or PCR, scientists were baffled by the observed accuracy of translation of mRNA into protein sequences – the so-called specificity paradox.
The observed accuracy is about one error per 10,000 amino acid residues, an astonishingly low rate. The sequence of amino acids in a protein is specified by pairing of triplets of nucleotides in messenger RNAs (codons) with triplets in transfer RNAs (anticodons)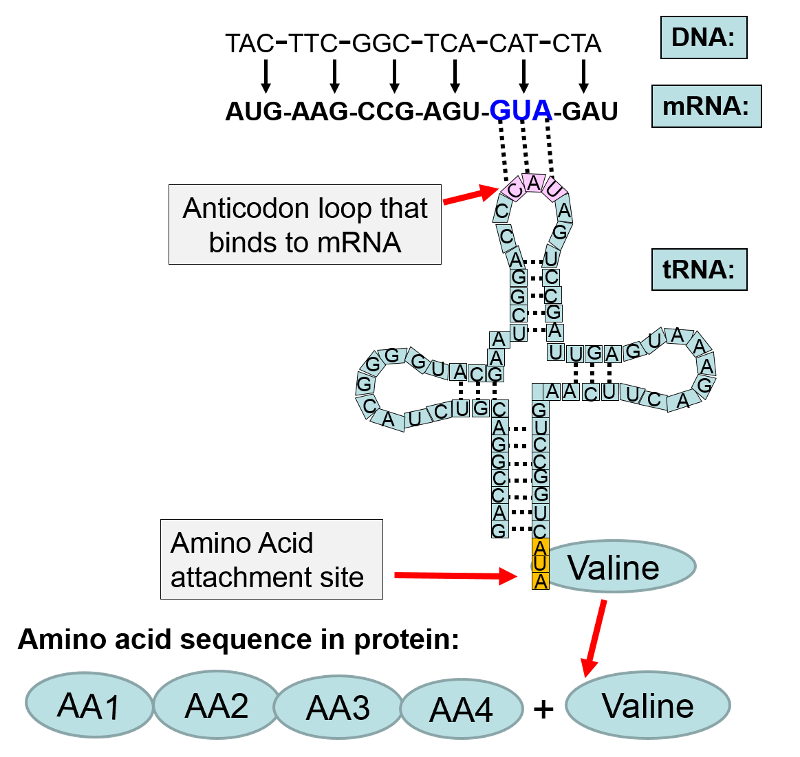
From DNA, Genetics and Evolution
Given the known tendency of RNAs to form mismatches, we’d expect the error rate in coding to be about 1 per 100 – a hundred times greater than the observed rate.
A number of explanations for the specificity paradox were put forth – conformational constraints, induced-fit models, manipulation of local charge distributions and dielectric constants. Although ribosomes do appear to take advantage of these mechanisms, they are insufficient to account for the observed specificity.
Molecular biologists generally think in pictures – we tend to believe evidence and mechanisms that are easily visualized. So it took a physicist (I’m sorry to say) to come up with the explanation: kinetic proofreading[1].
The essential insight of Hopfield (the physicist) was that equilibrium models such as induced fit would always fail because they were reversible – although they might reject some mismatches, they would introduce new ones, for no net gain. Hopfield proposed that adding an irreversible reaction after an initial scan would allow the scanning reaction to be repeated. Critically, this would square the intrinsic specificity of the scan – codon:anticodon specificity would go from 1 error in 100 to 1 in 10,000.
From Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Molecular biologists hate this stuff.
This idea met much resistance, but is widely accepted now[2] . It has been extended to a variety of high-accuracy biological processes, including DNA replication and antigen screening. Unfortunately, CRISPR is not one of these high-accuracy processes.
How safe does CRISPR have to be used on healthy people? Vaccines are a medicine that is given to healthy people. I’d say that a treatment to make someone taller or smarter would have to be at least as safe as vaccines to be acceptable.
Rates of serious injuries (not stuff like rashes, headaches or transient fevers) from most vaccines are so low that they are almost impossible to measure. A decent guess might be that 1 in 100,000 otherwise healthy people suffer serious health effects from vaccines[3].
We don’t know what the rate of injury from CRISPR editing is yet. But it appears that hundreds of new mutations are introduced into mouse embryos when treated with current technology[4] . Most of these will be neutral and have no real effect on health. But even if only 1 in 1000 mutations are harmful, that means that every tenth mouse is injured. That’s way too high.
There are of course efforts to improve the specificity of CRISPR. Some of them even invoke kinetic proofreading methods[5] and significantly improve accuracy. But they need to get at least a 1000-fold better before being used to treat healthy people.
That’s a lot, and we may never get there. CRISPR tech can still be used to treat cells outside the body (where they can be screened for defects) and to treat serious diseases inside the body (where the benefit outweighs the risk). But for “improving” otherwise healthy people? Unlikely.
Footnotes
[1] Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity.
[2] A recent intermezzo at the Ribosome Club
[4] CRISPR gene editing can cause hundreds of unintended mutations
[5] Enhanced proofreading governs CRISPR-Cas9 targeting accuracy.


